Joint Research Activity 3 (JRA3)
Building a consistent Information System in the different nodes and defining standardization strategies
Unification of information systems across infrastructures and countries is essential for an advanced community on phenotyping. This is to enable members to share data and to establish the open data strategy required in the call.
JRA3 aims at building and integrating a compatible, consistent information system that can be extended beyond EPPN2020. We design and provide methods and interfaces to manage, share, reuse and visualize heterogenous, high throughput plant phenotyping data stemming from different sources and/or in an interdisciplinary context. Another objective is to provide user friendly tools and a powerful environment beneficial to data analysis, integration and to the user communities.
|
Leader: Pascal Neveu (INRA Mistea) |
+33 4 99 61 24 12 |
|
|
Deputy Leader: Björn Usadel (FZJ) |
+49 2461 61-9503 |
Main activities
The first objective (TJRA1) is to ensure that data can be used across experiments and phenotyping installations. We developed methods that ensure that (i) environmental data are consistent across installations, with a common minimum list of environmental variables. We use the triple information rule: every measured variable (e.g. incident light) is characterized by (i) a precise definition (e.g. photosynthetic photon flux density), (ii) a method (sensors, positions) and (iii) a unit. Phenomic variables (e.g. plant leaf area or root elongation rate) are tracked with the same triplet, in such a way that any future user can exactly know what was measured.
The second objective (TJRA2) is to design an information system able to be deployed in a diversity of phenomic installations, including controlled conditions and field installation. A prototype has been released and tested.
The third objective (TJRA3) is to design a suite of methods allowing information systems at different sites to be ‘Interoperable’. For that, we are developing web services common to different European sites, so a user can access different datasets in different sites with a common query (e.g. ‘time course of leaf growth rate of a given genotype in all installations’).
The fourth objective (TJRA4) is to deploy and assess the distributed information system described above across European installations. We are installing information systems in installations who accepted to be beta testers for adapting ontologies, algorithms and methods to local systems and variables.
Levels for user access and for a European infrastructure of Phenomics
EPPN2020 has defined two levels for each activity. The first level allows the considered installation or local infrastructure to offer transnational accesses, the second is the objectives desirable for a future European infrastructure of phenomics, at installation/local infrastructure level. We define here a third level, at European infrastructure level.
Level 1: Data Identification and organization ('Findable' 'Reusable') Data are stored with backup or duplication service. Sensors are identified with persistent and non-ambiguous identifiers (e.g. URIs), allowing one to trace type/model, spatial positions and calibrations. Plants/pots/microplots are identified with URIs allowing one to trace spatial positions and characteristics. Environmental variables are based on ontologies allowing one to relate sensor outputs to time courses and/or spatial distribution of variables. Phenotypic variables are based with ontologies, methods and units. Events during experiments are traced with URIs using public ontologies (whenever possible). Datasets are organized in non-proprietary format with the necessary metadata.
Level 2: Local information system for environmental and phenotypic datasets ('Findable', 'Accessible', 'Reusable' at the level of a single local infrastructure). The local infrastructure installs an information system that allows connecting the information obtained in level 1. It will comply with domain standard such as MIAPPE specifications whenever relevant and provides rapid query of any combination of information (e.g. trait values for plants of a given genotype in a given range of environments, across experiments), and rapid detection of problems associated with an experiment (e.g. origin of outlier points or plants due to calibration issues or particular events). This is based on stabilized ontologies and provides linked data features (mainly based of semantic web methods).
Level 3: A multi local information system facilitating meta-analyses ('Findable', 'Accessible', ‘Reusable’, Interoperable' at the level of the European community ). This third step connects local infrastructures that have reached level 2. It allows any user to perform the same queries as in level 2, but at a multi local level.
An ontology-driven multi source and multi scale information system (PHIS)
An open-source information system (PHIS) was released and published (Neveu et al 2019 New Phytologist 221: 588–601) for plant phenotyping experiments in field and greenhouse installations. It non-ambiguously identifies all objects and traits in an experiment and establishes their relations via ontologies and semantics that apply to both field and controlled conditions. For instance, the genotype is declared for a plant or plot and is associated to all objects related to it. Events such as successive plant positions, anomalies and annotations are associated to objects so they can be easily retrieved. Its ontology-driven architecture is a powerful tool for integrating and managing data from multiple experiments and platforms, for creating relationships between objects and enriching datasets with knowledge and metadata. It interoperates with external resources via Web services, thereby allowing data integration into other systems, e.g. modelling platforms or genetic databases. It is progressively deployed in European local infrastructures, but also beyond Europe. Phis is publicly available at https://github.com/OpenSILEX under a GNU Affero General Public License version 2.
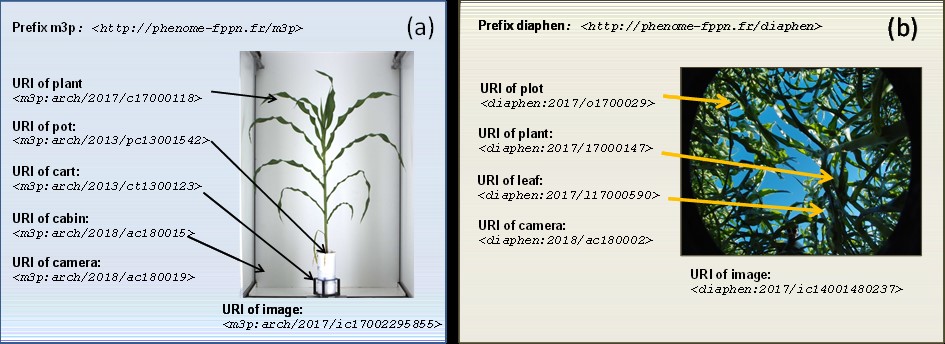
An example of the use of Unique Resource Identifiers (URIs) for identifying all objects present in single images taken in (a) greenhouse and (b) field experiments

